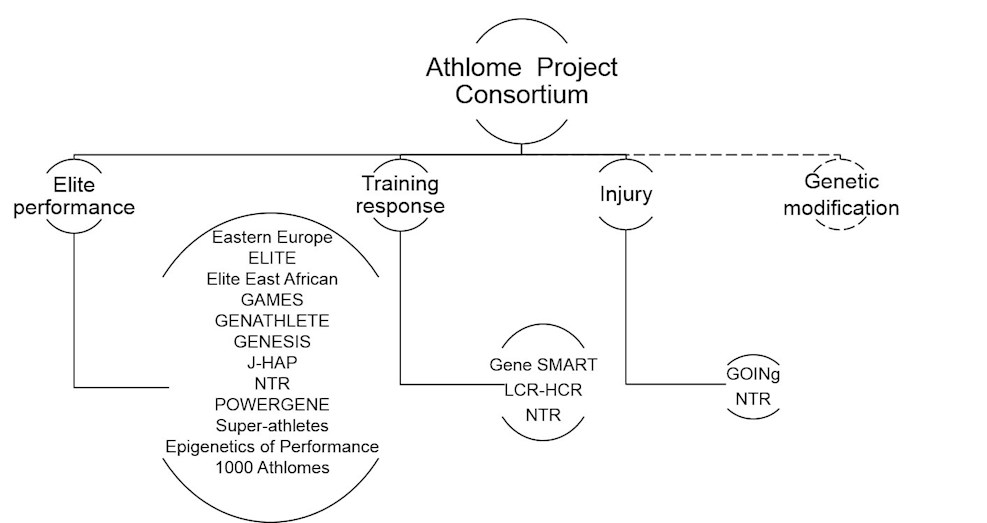
The 1000 Athlome Project aims to sequence 1000 genomes of sprinters and distance runners of West and East African descent. Phase 1 of the project has already commended and involves the sequencing of 12 sprinters and 12 distance runners of the highest level (i.e. world record holders, Olympians and World Champions). Phase 2 (2016-2018) will involve increasing the sample size for sequencing to 100 genomes. The pool of the runners to be sequenced will be expanded to 1000 by 2020 (Phase 3). An important aim of this sequencing project is to document the genotype distribution of elite east and west African athletes. The large amount of genotype data to be generated from the 1000 Athlome project will serve 1) as a reference panel for future performance studies, and 2) to guide other extreme phenotype studies in medical science.
Principal Investigators
- Masashi Tanaka (Tokyo Metropolitan Institute of Gerontology, JPN)
- Yannis Pitsiladis (University of Brighton, GBR)
The Russian and Belarusian cohorts, the Genetics and Epigenetics of Lithuanian Athletes from Kaunas (GELAK) and Vilnius (GELAV), and the Genome of Ukrainian Athletes Project (GUAP) have consolidated to identify genetic and epigenetic variations associated with high-level sports performance. The cohort comprises East Europeans (from Belarus, Lithuania, Russia, and Ukraine; in total n = 8,228 athletes and n = 4,121 controls). The athletes are grouped into international (including participants in Olympics and World Championships), national, regional, or local/non-competitive categories. These include biathletes, distance runners, cyclists, triathletes, kayakers, rowers, canoers, modern pentathletes, orienteers, skiers, speed skaters, short-trackers, walkers, weightlifters, bodybuilders, powerlifters, strongmen, sprint runners (≤ 400 m), sprint swimmers (50 - 100 m), decathletes, heptathletes, combat athletes, field athletes, bobsleigh athletes, rhythmic and artistic gymnasts, figure skaters, fencers and team ball-sport players. A portion of the participants have been evaluated with a variety of quantitative performance- and health-related assessments, including strength/power-related measurements, agility/speed-related measurements, balance, flexibility and coordination measurements, endurance-related measurement, skeletal muscle biopsy, and health-related measurements.
Principal Investigators
- Ildus I Ahmetov (Volga Region State Academy of Physical Culture, Sport and Tourism, RUS)
- Svitlana B Drozdovska (National University of Physical Education of Ukraine, UKR)
- Colin Moran (University of Stirling, GBR)
- Valentina Ginevičienė (Vilnius University, LTU)
- Andrei A Gilep (Institute of Bioorganic Chemistry NASB, BLR)
Funding
Eastern Europe population studies (The Russian and Belarusian cohorts, GELAK, GELAV, GUAP) were supported by the grants from the Federal Agency for Physical Culture and Sport of the Russian Federation and the Ministry of Education and Science of the Russian Federation (contract number 02.522.11.2004), the Federal Medical-Biological Agency of the Russian Federation (“Sportgen project”), Republic of Belarus (State program of development of physical culture and sports for 2011-2015) and Royal Society International Joint Project grant from the United Kingdom (code F-90014). GELAV (Epigenetics of Lithuanian Athletes from Vilnius) project was developed by the Lithuanian National Olympic Committee and Lithuanian Olympic Sports Centre, while actual research was carried out at the Vilnius University, where Lithuanian athletes DNA samples are stored. We would like to thank Prof. Vaidutis Kučinskas from the Department of Human and Medical Genetics, Faculty of Medicine, Vilnius University, Lithuania, for providing ideas and support for accessing the control samples.
Gallery
The Exercise at the Limit – Inherited Traits of Endurance (ELITE) consortium is a global initiative with the main objective to map the role that genetics plays in athletic ability versus environmental factors, such as training. Study participant (n > 500) selection is based on a physiological variable relevant for both health and sport performance, i.e., maximum oxygen uptake (V̇O2max). The main inclusion criterion is V̇O2max > 75 ml/kg/min for men and > 63 ml/kg/min for women, respectively. The consortium is continuously expanding and is recruiting athletes from all over the globe (with main focus on Caucasians, North East Africans, East Asians and South Americans) who are successful in endurance sports (running, cycling, cross country skiing, triathlon, rowing). Analyses currently include enhanced whole exome sequencing and GWAS (1.7 million SNPs). The combination of analytic methods will enable findings and differentiation between common variants with small effects and novel rare variants with larger effects. The aim is also to investigate differences between gender and ethnicity.
Principal Investigators
- Euan A Ashley
- C Mikael Mattsson
- Matthew Wheeler
- Daryl Waggott (Stanford University, USA)
Funding
ELITE is supported by SAP/Stanford Sequencing Initiative and Women’s Heart Health at Stanford.
The consortium also aims to study the East African running success by analyzing data from previously recruited subjects: (i) 76 endurance runners (64 men) and 38 sprint and power event athletes (18 men) from the Ethiopian national athletics teams, 315 controls from the general Ethiopian population (281 men), 93 controls from the Arsi region of Ethiopia (80 men) and (ii) 291 elite Kenyan endurance athletes (232 men) and 85 control participants (40 men). Seventy (59 men) Kenyan athletes had competed internationally and achieved outstanding success.
Principal Investigators
- Yannis Pitsiladis (University of Brighton, GBR)
- Robert Scott (University of Cambridge, GBR)
It is clear from animal and human studies that epigenetic marks play a role in the modulation of gene expression in relevant tissues. There also are indications that epigenetic marks can be altered by acute and chronic exercise in skeletal muscle and adipose tissue where they have been studied. Thus individual differences in any exercise-related traits can potentially be explained by not only the impact of DNA sequence variation on biology and behavior but also by the effects of epigenomic signaling on gene expression. We are formulating the hypothesis that elite athletic performance is influenced by epigenomic alterations, facilitating morphological, physiological, metabolic, cognitive, emotional and behavioral changes that empower the athlete to push performance beyond existing boundaries. We envisage testing this hypothesis by recruiting twin athletes competing at the Olympic or World Championship levels.
Principal Investigators
- Vassilis Klissouras (University of Athens, GRC)
- Yannis Pitsiladis (University of Brighton, GBR)
An international consortium (GAMES) was established to compare elite endurance athletes and ethnicity-matched controls in a case-control study design. GWASs were undertaken on two cohorts of elite endurance athletes and controls (GENATHLETE and Japanese endurance runners), from which a panel of 45 candidate SNPs was identified. These markers were tested for replication in seven additional cohorts of endurance athletes and controls: from Australia, Ethiopia, Japan, Kenya, Poland, Russia and Spain. The study is based on a total of 1,520 endurance athletes (835 of them had competed in World Championships or Olympic Games) and 2,760 controls.
Principal Investigators
- Claude Bouchard
- Tuomo Rankinen (Pennington Biomedical Research Centre, Louisiana State University, USA)
- Noriyuki Fuku (Juntendo University, JPN)
- Yannis Pitsiladis (University of Brighton, GBR)
- Bernd Wolfarth (Humboldt University, DEU)
- Alejandro Lucia (Universidad Europea de Madrid, SP)
Funding
GAMES was partially funded by the Prince Faisal Prize awarded to Drs C. Bouchard, T. Rankinen, M. Sarzynski and B. Wolfarth. CB is partially funded by the John W Barton Jr Chair in Genetics and Nutrition. The study in Russia was supported by a grant from the Federal Medical-Biological Agency (“Sportgen project”), http://fmbaros.ru/en/fmba/infor/. The Spanish group is funded by Cátedra Real-Madrid Universidad Europea, and Fondo de Investigaciones Sanitarias and Fondos Feder (grant # PI12/00914). This work in Japan was supported in part by grants from the programs Grants-in-Aid for Challenging Exploratory Research (24650414 to NF) from the Ministry of Education, Culture, Sports, Science and Technology; and by a grant-in-aid for scientific research from the Ministry of Health, Labor, and Welfare of Japan (to MM). No specific funding for this work was received for the studies performed in Australia, Poland, Kenya and Ethiopia.
The study was launched in 1993 with the aim of identifying DNA variants that are present at different frequencies between elite endurance athletes and sedentary controls. Male endurance athletes and controls were recruited from Canada, Finland, Germany and the USA. The cohort assembled to date includes 315 elite endurance athletes and 320 matched controls. Selection criteria for the all-male endurance athlete sample include that they had to be athletes of national or international caliber with a V̇O2max of at least 75 ml/kg/min. The mean value for the 315 athletes is currently 79 ml/kg/min while the mean for the 320 control subjects reached 40 ml/kg/min. Multiple candidate genes have been studied using the resources of GENATHLETE. A genome-wide screen for common variants has been performed on GENATHLETE and further studies are focusing on nuclear and mitochondrial DNA sequencing.
Principal Investigators
- Claude Bouchard
- Tuomo Rankinen (Pennington Biomedical Research Centre, Louisiana State University System, USA)
- Bernd Wolfarth (Department of Sports Medicine, Charite Medical School, Berlin, Germany)
- Louis Perusse (Laval University, Quebec, Canada)
- Rainer Rauramaa (University of Eastern Finland, Kuopio, Finland)
Gallery
The Gene SMART (Skeletal Muscle Adaptive Response to Training) study aims to identify the gene variants that predict the skeletal muscle response to both a single bout and 4 weeks of High-Intensity Interval Training (HIIT) in three different training centres. While the lead training and testing centre is located in Victoria University, Melbourne, two other centres have been launched at Bond University, Australia and the University of Sao Paulo, Brazil. The cohort is made up of moderately-trained, healthy participants (all men aged 20-45 years, body mass index ≤ 30 kg/m2). Participants are undergoing similar exercise testing and exercise training in three different laboratories. Dietary habits are assessed by questionnaire and nutritionist consultation. Activity history is assessed by questionnaire and current activity level is assessed by activity monitoring. A number of muscle and blood analyses are to be performed, including genotyping, mitochondrial respiration, transcriptomics, proteomics, and enzymes activity. Currently ~40 participants have finished the study and the overall aim is to train 250 participants. The Gene SMART also includes baseline and post-training testing and sampling for all participants before, during and after training. The participants serve as their own controls.
Principal Investigators
- David Bishop
- Nir Eynon (Victoria University, AUS)
Funding
Gene SMART Study is partly supported by Dr Eynon’s Australian Research Council Early career Fellowship (ARC DECRA) DE#140100864, and by the Victoria University Central Research Grant Scheme (CRGS).
The GENetics of Elite Status In Sport (GENESIS) consortium aims to identify molecular genetic characteristics associated with successful sports performance. The cohort (current n > 1,200) is mainly composed by UK athletes. Sports include marathon running and other track-and-field athletics, cycling and team sports (e.g., soccer). The RugbyGene Study is a major subcomponent of GENESIS and focuses on rugby (both union and league codes). Objectives of GENESIS are: (i) to increase current cohort size substantially; (ii) to apply hypothesis-free approaches to identify molecular genomic markers; (iii) to expand GENESIS from genomics to other “omics”; and (iv) to combine the “omics” data with athlete health and performance data to maximize practical impact of GENESIS.
Principal Investigators
- Alun G Williams
- Stephen H Day
- Georgina K Stebbings (Manchester Metropolitan University, GBR)
- Robert M Erskine (Liverpool John Moores University, GBR)
- Hugh E Montgomery (University College London, GBR)
Gallery
The recently established Genomics Of INjuries (GOINg) consortium aims to identify DNA sequence variants that modify the risk of anterior cruciate ligament (ACL) injuries. It is the only consortium within the ATHLOME Project to specifically investigate exercise-associated musculoskeletal injuries. The plan is to screen current known loci for ACL injury susceptibility in larger data sets in an attempt to determine if these loci remain as susceptibility loci across all populations using the hypothesis-driven candidate gene case-control study design. Care will be taken to use the same criteria to accurately phenotype, with respect to ancestry, sporting and occupational details, injury profile and mechanism(s) of injury, other injury history and family history, as well as, other appropriate medical history and medication use. The actual functional significance of the identified variants will also be investigated. This initial phase will be followed by the ACL sequencing project and the research objectives will be eventually expanded to include other “omics”. Thus far, ACL rupture consortium has collected DNA samples and clinical, as well as physical and occupational activity information from subjects from South Africa, Poland, Australia, Russia and Italy.
Principal Investigators
- Malcolm Collins
- Alison September
- Michael Posthumus (University of Cape Town, ZAF)
- Nir Eynon (Victoria University, AUS)
- Pawel Cieszczyk (University of Szczecin, POL)
Funding
GOINg is supported by funds from the National Research Foundation (NRF) of South Africa and the Thembakazi Trust.
Gallery
The Japanese Human Athlome Project (J-HAP) focuses on the study of genes associated with physical performance and its related phenotypes (e.g., muscle mass, muscle fibre type, V̇O2max). The cohort is comprised of Japanese athletes (currently > 2,400, mainly international and national levels) and healthy Japanese controls (currently > 1,000). These athletes are mainly track-and-field athletes and swimmers competing in endurance- and sprint/power-oriented events. Multiple “omics” approaches will be used to determine genes in talent identification in the Japanese population. Among the collected Japanese athletes’ and controls’ samples, approximately 200 muscle biopsies were obtained from both athletes and controls in order to investigate genetic variants associated with muscle fibre type distribution.
Primary Investigators
- Noriyuki Fuku (Juntendo University, JPN)
- Naoki Kikuchi (Nippon Sport Science University, JPN)
- Eri Miyamoto-Mikami (The National Institute of Fitness and Sports in Kanoya, JPN)
Funding
J-HAP project was supported by JSPS KAKENHI (Grant Numbers 21680050, 11J04771, 24650414, 26882041, 15H03081, and 15K16467) and by MEXT- commissioned projects.
The Netherlands Twin Register (NTR) is a population-based cohort recruiting both newborn and adult multiples and their family members with continuous longitudinal data collection. In the past 25+ years, around 40% of all twins and multiples in the Netherlands have taken part in research projects of the NTR. Family members and spouses of twins also took part, leading to a total of over 185,000 participants across multiple research projects. The longitudinal information that has been collected extends from genotype to biomarkers, gene expression to rich behavioral information including biennial reports on (competitive) sports participation and performance level and on injuries related to sports. In its sports research track, NTR aims to understand the interplay between genetic and environmental factors shaping individual differences in sports participation and performance. In the NTR, participants are recruited as newborns and followed into young adulthood, 520 have played competitively at a regional and 189 at a national level. Main sports that Dutch adolescents/young adults engage in are swimming, tennis, bicycling, soccer and field hockey. The longitudinal data collection of the NTR is ongoing and securely funded for the next 5 years.
Principal Investigators
- Eco de Geus
- Meike Bartels (VU University and VU medical centre, NLD)
Funding
NTR’s funding was obtained from the Netherlands Organization for Scientific Research (NWO) and The Netherlands Organisation for Health Research and Development (ZonMW) grants 904-61-090, 985-10-002, 912-10-020, 904-61-193, 480-04-004, 463-06-001, 451-04-034, 400-05-717, Addiction-31160008, Middelgroot-911-09-032, Spinozapremie 56-464-14192, Center for Medical Systems Biology (CSMB, NWO Genomics), NBIC/BioAssist/RK(2008.024), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI –NL, 184.021.007). VU University’s Institute for Health and Care Research (EMGO+ ) and Neuroscience Campus Amsterdam (NCA); the European Science Foundation (ESF, EU/QLRT-2001-01254), the European Community’s Seventh Framework Program (FP7/2007-2013), ENGAGE (HEALTH-F4-2007-201413); the European Research Council (ERC Advanced, 230374, ERC Starting grant 284 167), Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06), the Avera Institute, Sioux Falls, South Dakota (USA) and the National Institutes of Health (NIH, R01D0042157-01A, MH081802; R01 DK092127-04, Grand Opportunity grants 1RC2 MH089951 and1RC2 MH089995). Computing was supported by BiG Grid, the Dutch e-Science Grid, which is financially supported by NWO.
Gallery
The POWERGENE consortium aims to characterise the elite sprint/power athlete genotype. The internationally competitive (Olympic/Word championship qualifiers) sprint/power athletes are from: Australia, Belgium, Greece, Italy, Jamaica, Japan, Lithuania, Poland, Spain, the U.S.A., Brazil (replication cohort), and Russia (replication cohort). They will be compared with sub-elite athletes (national qualifiers), endurance athletes, team athletes and controls. The current cohort consists of female (n = 264) and male (n = 481) specialist power athletes across three major ethnicities (i.e., West African and East Asian ancestries, and Caucasians). Sprint/power athletes include those individuals competing in track (≤ 800 m) and field (jump, throw) events, cycling (track), swimming (≤ 200 m), gymnastics (artistic), weightlifting, judo, speed-skating and power lifting. Endurance athletes (n = 586) include track and road running specialists (> 800 m), rowers, cyclists, swimmers (> 200 m), triathletes and ironmen. Team sports (n = 862) include football (soccer), cricket, hockey, volleyball and basketball.
Principal Investigators
- Yannis Pitsiladis (University of Brighton, GBR)
- Kathryn North (Murdoch Childrens Research Institute, AUS)
- Nir Eynon (Victoria University, AUS)
Funding
POWERGENE GWAS genotyping in Jamaicans, African-Americans (the U.S.A. cohort) and Japanese was funded by JSPS KAKENHI (Grant Number 21680050 and 24650414).
Gallery
The purpose of the Low Capacity Rats-High Capacity Rats (LCR-HCR) model is to serve as a resource for the in-depth study of rat models to resolve the extremes of exercise and health. By connecting clinical observation with a theoretical base, the working hypothesis is that: variation in capacity for energy transfer is the central mechanistic determinant between disease and health (energy transfer hypothesis). As an unbiased test of this hypothesis, this study showed that two-way artificial selective breeding of rats for low and high intrinsic endurance exercise capacity also produced rats that differed for numerous disease risks, including the metabolic syndrome, premature aging, fatty liver disease, obesity, and Alzheimer’s disease. Exercise capacity is a result of intrinsic capacity plus adaptation to all aspects of physical activity. To capture this biology, rats for low and high response to 8 weeks of treadmill running exercise were selectively bred. Thus, the study has models that represent the 4 “corners” of exercise capacity. These contrasting animal model systems may prove to be translationally superior relative to more widely used simplistic models for understanding disease conditions. The rat models may be deeply explored to discover causal mechanisms and develop effective therapeutics. These rats are being studied at over 50 institutions in 11 countries.
Principal Investigators
- Steven Britton
- Lauren Koch (University of Michigan, USA)
Funding
Rat models of exercise and health (LCR-HCR rat model) was funded by the Office of Research Infrastructure Programs/OD grant R24OD010950 and by grant R01DK099034 (to LGK and SLB) from the National Institutes of Health. We acknowledge the expert care of the rat colony provided by Molly Kalahar and Lori Heckenkamp. Contact: LGK lgkoch@umich.edu or SLB brittons@umich.edu for information on the LCR and HCR rats: these rat models are maintained as an international resource with support from the Department of Anesthesiology at the University of Michigan, Ann Arbor, Michigan.
Gallery
The study aims to identify genetic variants associated with elite athletic performance (power versus endurance) and to study potential ethnic differences in this predisposition. A further aim is to study the functional significance of the identified variants. A GWAS will be carried out in 3,000 consented elite athletes who tested negative for doping substances at the Anti-Doping Laboratories, Federazione Medico Sportiva Italiana (FMSI) and Anti-Doping Lab Qatar (ADLQ), using Illumina genotyping technologies. Comparing genotype frequency distribution of elite athletes from European countries (where most of FMSI samples will be obtained) against those from South Asian and African countries (where most of ADLQ samples are expected to be obtained) would help to identify potential ethnic differences in the genetic predisposition to athletic performance. Subsequently, urine metabolome in a subset of these athletes (1,000 subjects) will be performed, and will be related to the athlete’s sporting discipline.
Principal Investigators
- Mohamed El-Rayess
- Costas Georgakopoulos
- Mohammed Alsayrafi (ADLQ, QAT)
- Francesco Botre (FMSI, ITA)
- Karsten Suhre (Weill Cornell Medical College in Qatar, QAT)
- Mike Hubank (University College London, GBR)
Funding
Super-athletes: Genes and Sweat is partly funded by Qatar National Research Foundation NPRP grant (NPRP 7-272-1-041).





















![IMG_20160209_111455481_HDR[1].jpeg](/umbraco/imagegen.ashx?width=100&height=100&image=/media/1042/img_20160209_111455481_hdr-1.jpeg)

![IMG_20160209_111750247[1].jpeg](/umbraco/imagegen.ashx?width=100&height=100&image=/media/1043/img_20160209_111750247-1.jpeg)
![IMG_20160209_113025054_HDR[1].jpeg](/umbraco/imagegen.ashx?width=100&height=100&image=/media/1045/img_20160209_113025054_hdr-1.jpeg)
![IMG_20160209_113030181[1].jpeg](/umbraco/imagegen.ashx?width=100&height=100&image=/media/1046/img_20160209_113030181-1.jpeg)
![IMG_20160209_113100814_HDR[1].jpeg](/umbraco/imagegen.ashx?width=100&height=100&image=/media/1047/img_20160209_113100814_hdr-1.jpeg)
![IMG_20160209_115817400[1].jpeg](/umbraco/imagegen.ashx?width=100&height=100&image=/media/1048/img_20160209_115817400-1.jpeg)
![IMG_20160209_115846868[1].jpeg](/umbraco/imagegen.ashx?width=100&height=100&image=/media/1049/img_20160209_115846868-1.jpeg)











